I recently came across the German Tank Problem in the docs for Sampyl, a fairly new Python library for Markov Chain Monte Carlo sampling. This problem is also an exercise in "Think Bayes" by Allen Downey, although there it was called "the locomotive problem."
Here's the problem statement:

Suppose four tanks are captured with the serial numbers 10, 256, 202, and 97. Assuming that each tank is numbered in sequence as they are built, how many tanks are there in total?
The problem is named after the Allies' use of applied statistics in World War II to analyze German production capacity for Panzer tanks, and while this statement of it is hugely simplified, the actual story of how that turned out is fascinating:
After the war, the allies captured German production records, showing that the true number of tanks produced in those three years was [...] almost exactly what the statisticians had calculated, and less than one fifth of what standard intelligence had thought likely.
Problem set-up🔗
We start with a simplified story about how the data is generated — the generative model — and then attempt to infer specifics about that story (the parameters) based on what data we ended up with. In other words, we're constructing a simple model of how the universe could be arranged in such a way that we ended up seeing these four numbers, because we assume that random chance was involved and things could have turned out differently.
Here's our model:
Choose a number of tanks
Of those
Of course, we actually start with the data
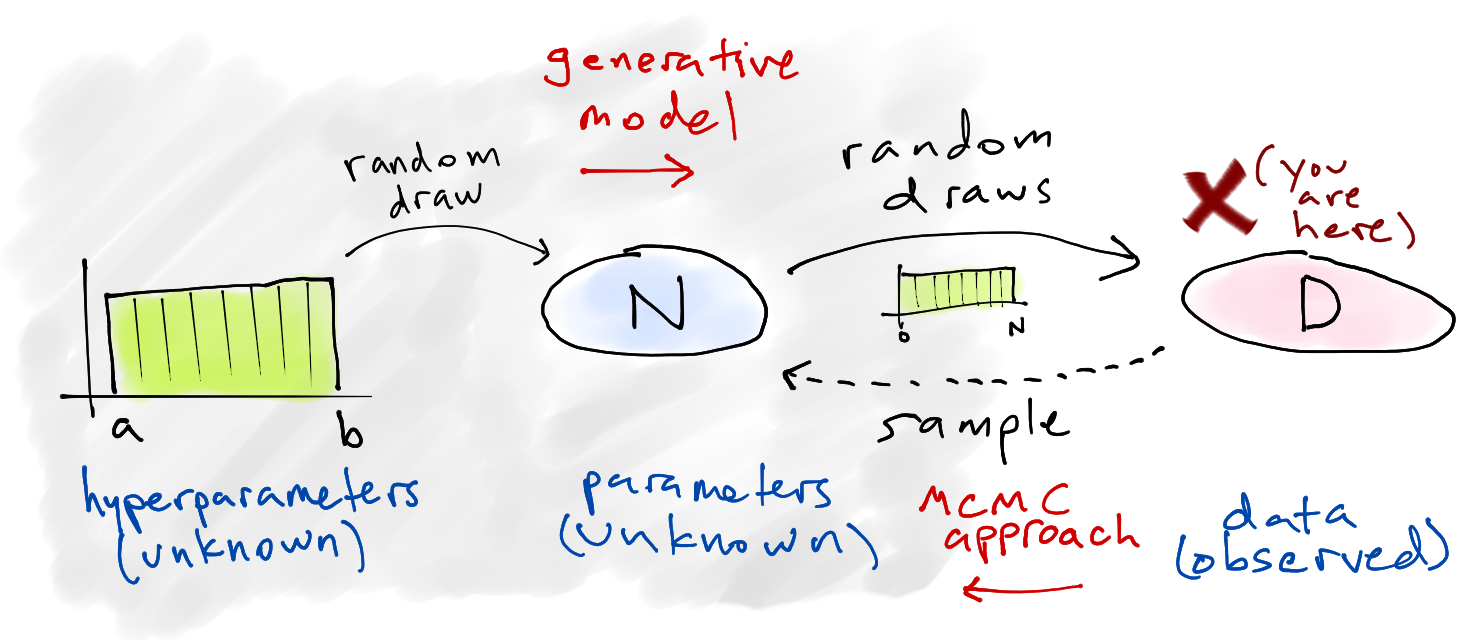
Solving it🔗
The Wikipedia article explains both the frequentist and Bayesian approaches, and the example in the Sampyl docs does a good job of explaining the setup for the Bayesian version. Here's the gist: we want to know the posterior probability of the number of tanks
The right-hand side breaks down into two parts. The likelihood of observing all the serial numbers we saw is the product of the individual probabilities
The prior over
The MCMC strategy will be to try a bunch of different values for
PyMC3🔗
PyMC seems to be most one of the most commonly used libraries for MCMC modeling in Python, and PyMC3 is the new version (still in beta). The tutorial in the project docs is a good read in and of itself, and Bayesian Methods for Hackers uses its predecessor PyMC2 extensively.
import numpy as np
import pymc3 as pm
# D: the data
y = np.array([10, 256, 202, 97])
model = pm.Model()
with model:
# prior - P(N): N ~ uniform(max(y), 10000)
# note: we use a large-ish number for the upper bound
N = pm.DiscreteUniform("N", lower=y.max(), upper=10000)
# likelihood - P(D|N): y ~ uniform(0, N)
y_obs = pm.DiscreteUniform("y_obs", lower=0, upper=N, observed=y)
# choose the sampling method - we have to use Metropolis-Hastings because
# the variables are discrete
step = pm.Metropolis()
# we'll use four chains, and parallelize to four cores
start = {"N": y.max()} # the highest number is a reasonable starting point
trace = pm.sample(100000, step, start, chain=4, njobs=4)
# summarize the trace
pm.summary(trace)
%matplotlib inline
from matplotlib import pyplot as plt
# plot the trace
burn_in = 10000 # throw away the first 10,000 samples
pm.traceplot(trace[burn_in:])
plt.show()
So the mean here was 384, and the median was 322.
PyStan🔗

PyStan "provides an interface to Stan, a package for Bayesian inference using the No-U-Turn sampler, a variant of Hamiltonian Monte Carlo." Stan, the underlying package, is designed to be a successor to JAGS, BUGS, and other hierarchical modeling tools.
Here's the same model implemented with PyStan. Two notes about how this differs:
The model specification is just a string written in the Stan sytax. PyStan is a thin wrapper that just handles sending off the model for compilation along with the data and then bringing back the results — it is not an attempt to write models in Python.
Stan doesn't currently have a convenient way to handle discrete parameters, which seems to be because the No-U-Turn Sampler (NUTS) and Hamiltonian Monte Carlo are designed for continuous variables only. For that reason, this formulation is for the continuous version of the problem and uses uniform instead of discrete uniform.
import pystan
ocode = """
data {
int<lower=1> M; // number of serial numbers observed
real y[M]; // serial numbers
}
parameters {
real<lower=max(y)> N;
}
model {
N ~ uniform(max(y), 10000); // P(N)
y ~ uniform(0, N); // P(D|N)
}
"""
data = {'y': y, 'M': len(y)}
fit = pystan.stan(model_code=ocode, data=data, init='init_r', iter=100000, warmup=1000, chains=4)
fit
fig = fit.plot()
fig.set_size_inches((12, 2))
plt.tight_layout()
plt.show()
fit.extract()['N'].mean()
The mean was again 384 and the median was 323, closely matching the results from above as expected.